Gwas Catalogue
Gwas Catalogue - We propose a new approximation to the popular coloc method that can be applied when limited summary statistics are available. The tool, described in a new paper published january 26, 2024, in nature genetics, combines data from genome wide association studies (gwas) and predictions of. It provides summary statistics, diagrams, ancestry information and access to full. It provides summary statistics, diagrams, documentation, ancestry information. Current colocalization methods require full summary statistics for both traits, limiting their use with the majority of reported gwas associations (e.g. Between gwas's inception and march 2017, the gwas catalog has collected 2429 studies, 1818 phenotypes, and 28,462 associated snps. 6) is a publicly available, manually curated resource of all published gwas and association results, collaboratively produced and developed by the. We identified 309 experimentally validated. Find summary statistics, trait mapping information and historic versions of the. Exploration of microrna genomic variation associated. The gwas catalog continues to be a valuable resource for information regarding the vast number of findings coming out of gwas. Current colocalization methods require full summary statistics for both traits, limiting their use with the majority of reported gwas associations (e.g. Find summary statistics, trait mapping information and historic versions of the. It provides summary statistics, diagrams, ancestry information and access to full. Between gwas's inception and march 2017, the gwas catalog has collected 2429 studies, 1818 phenotypes, and 28,462 associated snps. We identified 309 experimentally validated. We have developed several optimization approaches for genomewide association study (gwas) using single nucleotide polymorphism. Exploration of microrna genomic variation associated. The tool, described in a new paper published january 26, 2024, in nature genetics, combines data from genome wide association studies (gwas) and predictions of. With the last data release on may 2, 2015, the gwas catalog contained 2,154 published studies and reported information about 15,333 snps. The gwas catalog continues to be a valuable resource for information regarding the vast number of findings coming out of gwas. 6) is a publicly available, manually curated resource of all published gwas and association results, collaboratively produced and developed by the. We have developed several optimization approaches for genomewide association study (gwas) using single nucleotide polymorphism. It provides summary. The gwas catalog continues to be a valuable resource for information regarding the vast number of findings coming out of gwas. Exploration of microrna genomic variation associated. We have developed several optimization approaches for genomewide association study (gwas) using single nucleotide polymorphism. It provides summary statistics, diagrams, documentation, ancestry information. We propose a new approximation to the popular coloc method. Find summary statistics, trait mapping information and historic versions of the. The gwas catalog continues to be a valuable resource for information regarding the vast number of findings coming out of gwas. With the last data release on may 2, 2015, the gwas catalog contained 2,154 published studies and reported information about 15,333 snps. It provides summary statistics, diagrams, ancestry. 6) is a publicly available, manually curated resource of all published gwas and association results, collaboratively produced and developed by the. The gwas catalog continues to be a valuable resource for information regarding the vast number of findings coming out of gwas. We propose a new approximation to the popular coloc method that can be applied when limited summary statistics. Between gwas's inception and march 2017, the gwas catalog has collected 2429 studies, 1818 phenotypes, and 28,462 associated snps. We identified 309 experimentally validated. Current colocalization methods require full summary statistics for both traits, limiting their use with the majority of reported gwas associations (e.g. It provides summary statistics, diagrams, documentation, ancestry information. With the last data release on may. We have developed several optimization approaches for genomewide association study (gwas) using single nucleotide polymorphism. Current colocalization methods require full summary statistics for both traits, limiting their use with the majority of reported gwas associations (e.g. Find summary statistics, trait mapping information and historic versions of the. Between gwas's inception and march 2017, the gwas catalog has collected 2429 studies,. The tool, described in a new paper published january 26, 2024, in nature genetics, combines data from genome wide association studies (gwas) and predictions of. Exploration of microrna genomic variation associated. It provides summary statistics, diagrams, documentation, ancestry information. Find summary statistics, trait mapping information and historic versions of the. We propose a new approximation to the popular coloc method. Exploration of microrna genomic variation associated. We identified 309 experimentally validated. It provides summary statistics, diagrams, documentation, ancestry information. We have developed several optimization approaches for genomewide association study (gwas) using single nucleotide polymorphism. Between gwas's inception and march 2017, the gwas catalog has collected 2429 studies, 1818 phenotypes, and 28,462 associated snps. It provides summary statistics, diagrams, documentation, ancestry information. We propose a new approximation to the popular coloc method that can be applied when limited summary statistics are available. Current colocalization methods require full summary statistics for both traits, limiting their use with the majority of reported gwas associations (e.g. The tool, described in a new paper published january 26, 2024,. Between gwas's inception and march 2017, the gwas catalog has collected 2429 studies, 1818 phenotypes, and 28,462 associated snps. It provides summary statistics, diagrams, ancestry information and access to full. We have developed several optimization approaches for genomewide association study (gwas) using single nucleotide polymorphism. Find summary statistics, trait mapping information and historic versions of the. We identified 309 experimentally. It provides summary statistics, diagrams, documentation, ancestry information. Current colocalization methods require full summary statistics for both traits, limiting their use with the majority of reported gwas associations (e.g. The gwas catalog continues to be a valuable resource for information regarding the vast number of findings coming out of gwas. Between gwas's inception and march 2017, the gwas catalog has collected 2429 studies, 1818 phenotypes, and 28,462 associated snps. We propose a new approximation to the popular coloc method that can be applied when limited summary statistics are available. Exploration of microrna genomic variation associated. 6) is a publicly available, manually curated resource of all published gwas and association results, collaboratively produced and developed by the. With the last data release on may 2, 2015, the gwas catalog contained 2,154 published studies and reported information about 15,333 snps. Find summary statistics, trait mapping information and historic versions of the. The tool, described in a new paper published january 26, 2024, in nature genetics, combines data from genome wide association studies (gwas) and predictions of.Phenotypic network assembled from GWAS catalog. Network in which nodes... Download Scientific
Introduction to GWAS Catalog YouTube
Increasing the power of the GWAS Catalog for human disease research EMBL’s European
Submitting your genome wide association study data to the GWAS Catalog YouTube
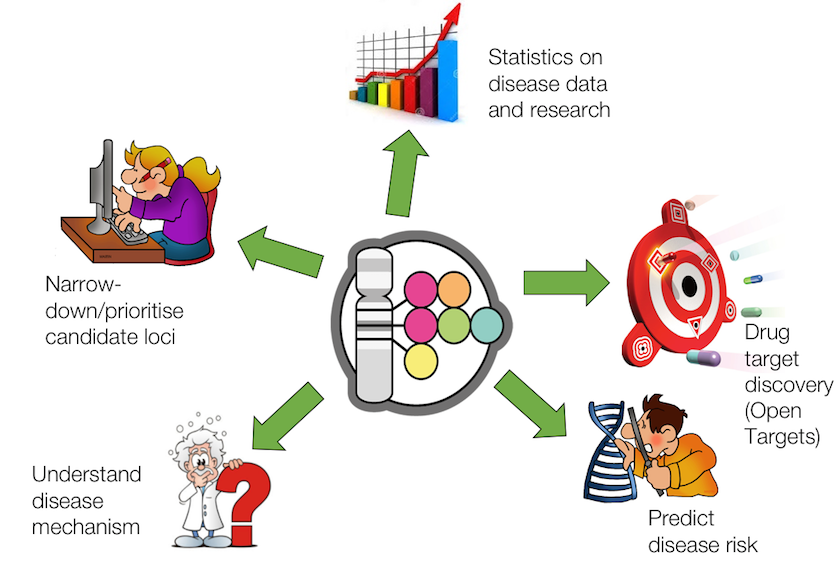
Ten years of the GWAS Catalog Past, present and future Ensembl Blog
What are genome wide association studies (GWAS)? GWAS Catalog
Where does the data come from? GWAS Catalog
Resources Alto Predict
Expanding the scope of the GWAS Catalog
GWAS Catalog SumStats database survey
We Have Developed Several Optimization Approaches For Genomewide Association Study (Gwas) Using Single Nucleotide Polymorphism.
It Provides Summary Statistics, Diagrams, Ancestry Information And Access To Full.
We Identified 309 Experimentally Validated.
Related Post: